Cancer Genetics and Genomics
 The Cancer Genetics and Genomics (CGG) Program seeks to better understand the cancer genome and leverage novel insights into cancer genomics to improve outcomes for cancer patients. The three main research themes of the CGG Program include: (1) invention of cancer genome and transcriptome profiling technology; (2) development of computational tools and resources for interpreting cancer genomes and transcriptomes; and (3) implementation of high-throughput genetic-screening technologies to comprehensively map cancer dependencies.
The Cancer Genetics and Genomics (CGG) Program seeks to better understand the cancer genome and leverage novel insights into cancer genomics to improve outcomes for cancer patients. The three main research themes of the CGG Program include: (1) invention of cancer genome and transcriptome profiling technology; (2) development of computational tools and resources for interpreting cancer genomes and transcriptomes; and (3) implementation of high-throughput genetic-screening technologies to comprehensively map cancer dependencies.
Program Co-leaders
Tobias Janowitz, M.D., Ph.D.
Adam Siepel, Ph.D.
![]() The Cancer Genetics and Genomics Program seeks to advance our understanding of the cancer genome through genetic, technological, and computational innovations. In fitting with the mission of the Cold Spring Harbor Laboratory (CSHL) Cancer Center as an NCI-designated Basic Laboratory Cancer Center, the Program focuses on fundamental research to better understand the genetic alterations that drive cancer development and progression. The research themes in the CGG Program are built on the premise that a deeper characterization of the cancer genome will improve the survival and quality of life for patients with cancer, and Program members are translating insights made about the cancer genome into novel diagnostic and prognostic biomarkers. In addition, the Program maintains a focus on the development of new genomic technologies and computational tools, which are being used to analyze the cancer genome, aid in cancer diagnosis, and discover novel cancer dependencies.
The Cancer Genetics and Genomics Program seeks to advance our understanding of the cancer genome through genetic, technological, and computational innovations. In fitting with the mission of the Cold Spring Harbor Laboratory (CSHL) Cancer Center as an NCI-designated Basic Laboratory Cancer Center, the Program focuses on fundamental research to better understand the genetic alterations that drive cancer development and progression. The research themes in the CGG Program are built on the premise that a deeper characterization of the cancer genome will improve the survival and quality of life for patients with cancer, and Program members are translating insights made about the cancer genome into novel diagnostic and prognostic biomarkers. In addition, the Program maintains a focus on the development of new genomic technologies and computational tools, which are being used to analyze the cancer genome, aid in cancer diagnosis, and discover novel cancer dependencies.
Over the last five years, several Program investigators have developed technologies for profiling cancer genomes and transcriptomes. These include diverse methods for in situ sequencing of RNA in tumors, methods to find structural variants, and single-cell analysis methods. Members are also developing computational tools for genome and transcriptome analysis, which reflects the interdisciplinary approach of members with expertise in both traditional bench research and computational biology. In addition, Program members are using high-throughput genetic-screening approaches for therapeutic target discovery and validation. This theme makes use of multiplexed screening technologies, pioneered by Program members, for high-throughput assessment of gene functions. To support these efforts, Program members are developing computational algorithms for interpreting the complex datasets generated through these studies. Several Cancer Center Shared Resources are pivotal for the discovery research in this Program, including the Flow Cytometry, Microscopy, and Sequencing Technologies & Analysis Shared Resources.
CSHL’s Thomas Gingeras awarded $2 million NSF grant
April 3, 2024
Climate change threatens crops with acidic soils and aluminum toxicity. Gingeras leads an international team tackling this problem head-on.
From plant genomics to a bioscience revolution
March 25, 2024
CSHL played a lead role in mapping the first plant genome. Today, that breakthrough fuels a whole new understanding of life on Earth.
A quiz for the ages
January 29, 2024
Want to know the secret to a long life? So do CSHL scientists. Take this short quiz to see what they’ve found out about aging and longevity.
From cancer genetics to cancer treatments
January 18, 2024
One cancer gene, one cancer genome, two Cold Spring Harbor Laboratory discoveries that helped shape the face of modern cancer medicine.
You say genome editing, I say natural mutation
October 19, 2023
CSHL scientists have discovered that evolution and genome editing in crops are less predictable than previously thought.
Holy immunity! Bat genes key against COVID, cancer
October 16, 2023
Rapid evolution has streamlined bats’ immune systems. This may explain why they’re resistant to cancer and viruses like Ebola or COVID-19.
A full-circle moment in cancer research
August 16, 2023
High-tech experiments by CSHL Postdoctoral Researcher Asad Lakhani and colleagues allow us to see early 20th-century hypotheses in a whole new light.
Genome hack reignites century-old cancer debate
August 14, 2023
ReDACT, a new genome-editing technique invented at Cold Spring Harbor Laboratory, could bring cancer research full circle.
The evolution of autism research
May 25, 2023
The conversation around autism has evolved over the past two decades. So has CSHL research. This retrospective shows how we’ve helped move the needle.
Autism in the family tree
May 23, 2023
CSHL scientists have studied the genetics of autism across hundreds of family trees. This animated video shows what they’ve found.
Siblings with autism share more of dad’s genome, not mom’s
May 22, 2023
CSHL study of more than 6,000 volunteer families overturns a long-held assumption about the genetic origins of autism spectrum disorder.
The CSHL School of Biological Sciences’ class of 2023
May 7, 2023
The School of Biological Sciences awarded 11 Ph.D. degrees this year. Here, the graduates reflect on their time and experiences at CSHL.
How well do you know autism spectrum disorders?
April 24, 2023
April is National Autism Awareness Month. Test your knowledge of autism spectrum disorders with this short quiz.
Genome analysis just got personal
March 30, 2023
A new algorithm created by CSHL researchers can help predict the health effects of millions of genetic variants found within a single person’s genome.
How evolved is your knowledge?
January 26, 2023
Test your knowledge of evolution with this quiz, inspired by the March 2023 performances of Isabella Rossellini’s play, Darwin’s Smile, at CSHL.
Unlocking cancer’s ancestry
December 27, 2022
New software may help reveal the complete connections between ancestry and cancer, which could lead to better, more personalized treatments.
CSHL professors on the cutting edge of scientific research
November 21, 2022
Four CSHL researchers were ranked among the world’s most cited in 2022.
Exposing the evolutionary weak spots of the human genome
September 22, 2022
Researchers built a computer program that tracks harmful mutations throughout human evolution. It may help uncover the origins of genetic diseases.
The genetic landscape of autism
August 16, 2022
CSHL Professor Michael Wigler discusses 20 years of research to paint a complete picture of the genetic causes of autism.
After 100 years of research, autism remains a puzzle
August 4, 2022
One geneticist is determined to piece together the causes of autism.
President’s essay: Foundations for the future
May 25, 2022
Strategically designed to spark scientific exchange and inspiration, CSHL is a unique research and education environment for advancing science.
Getting a step ahead of TB’s drug resistance evolution
February 15, 2022
Mutations are not random, with some kinds of changes occurring more often than others. CSHL researchers may be able to predict which direction evolution is li
Calculating the path of cancer
October 4, 2021
A new mathematical approach is helping cancer researchers at CSHL determine how mutations lead to different behaviors in cancerous cells.
CSHL SBS graduation: Ceremony canceled, graduates undaunted
August 23, 2021
In August 2021, the CSHL School of Biological Sciences awarded 7 Doctor of Philosophy degrees and two honorary degrees.
CSHL Ph.D. program: Graduating class of 2021
August 22, 2021
The CSHL School of Biological Sciences awarded Ph.D. degrees to seven students this year, who describe some of their experiences.
Cancer cells pick and choose chromosomes to survive
August 18, 2021
Cancer cells get rid of excess genetic baggage and double up on genetic protections to resist anti-tumor therapies.
On the wrong side of cell division
August 13, 2021
Bad cell divisions can lead to resistant tumors. Cancer cells benefit from extra drug resistance gene copies while dumping genes that could hurt them.
How to outwit evolution
July 21, 2021
CSHL Assistant Professor David McCandlish uses statistical methods to predict the evolution of antibiotic resistance in bacteria.
Research matters
June 8, 2021
Innovative research and educational activities never stopped during the COVID-19 pandemic.
How an antiviral drug can flatten the curve…in your cells
April 9, 2021
Scientists figured out how an antiviral drug in clinical trials combats COVID-19 and it’s better than expected.
CSHL wins ACS grant based on crowdsourced support
February 4, 2021
CSHL Fellow Jason Sheltzer received a grant through the American Cancer Society’s new social media platform TheoryLab™.
Birds of a feather do flock together
November 17, 2020
Researchers found a genetic mechanism for how brand new species acquire distinct traits.
Women’s Partnership luncheon raises $150,000 for research
September 29, 2020
The nineteenth annual Women’s Partnership for Science luncheon was held with social distancing to support CSHL women researchers.
Fuerth wins BBRF Young Investigator Grant in neuro research
September 25, 2020
Daniel Fuerth, a postdoctoral fellow in Assistant Professor Je Lee’s lab,will study the transfer of genetic material between neurons in the brain.
New genetic research to understand racial disparity in cancers
September 8, 2020
Cold Spring Harbor Laboratory will study the genetic contributions of ethnicity to colon, endometrial, and pancreas cancers in African Americans.
Why we’re a lot better at fighting cancer than we realized
August 12, 2020
Using data mining techniques, doctors have discovered dozens of anti-tumor drugs hiding in plain sight.
ENCODE3: Interpreting the human and mouse genomes
July 29, 2020
Researchers report on 900,000 regulatory elements in our genomes that could influence our health.
Smoking increases SARS-CoV-2 receptors in the lung
May 18, 2020
An increase in lung ACE2 may explain why smokers are particularly vulnerable to COVID-19.
Tuveson and Wigler elected AACR Academy Fellows
May 12, 2020
CSHL Cancer Center Director David Tuveson and Professor Michael Wigler were chosen as 2020 Fellows of the AACR Academy.
Dog adaptations are like brain adaptations
May 6, 2020
To understand autism, CSHL Professor Michael Wigler thinks about how biological systems adapt over generations.
The status of autism research—in verse
April 29, 2020
CSHL Professor Michael Wigler drafted a poem explaining what we know and what we need to learn about autism.
Predicting the evolution of genetic mutations
April 14, 2020
CSHL quantitative biologists have designed a computational approach for predicting the evolution of a rapidly mutating virus or cancer.
A science career path: David McCandlish
April 10, 2020
Assistant Professor David McCandlish is a quantitative biologist who walks the line between advanced mathematics and the life sciences at CSHL.
Carol Greider: Nobel Prize-winning rogue biologist
March 31, 2020
CSHL alumna Carol Greider won the Nobel Prize and is a champion for diversity. But her dyslexia almost derailed her career before it began.
New faculty Jeff Boyd studies breast cancer genomics
March 26, 2020
Professor Jeff Boyd joins the CSHL faculty, studying the growth and spread of breast cancer.
Extra chromosomes in cancers can be good or bad
February 24, 2020
Extra chromosomes are typical in cancerous tumor cells, but not all extra copies promote cancer growth.
The non-human living inside of you
January 9, 2020
A large part of human DNA doesn’t aid the normal workings of the body. This “junk DNA” contains ancient viruses that may spur diseases like ALS.
Ann Lin named in Forbes 30 Under 30 list for 2020
December 16, 2019
Ann Lin, a former intern in CSHL Fellow Jason Sheltzer’s lab, has been named a top entrepreneur on the Forbes 30 Under 30 list.
Bridge to education
December 15, 2019
CSHL’s DNA Learning Center builds new bridges between unique science education and diverse groups.
CSHL investigators rank among world’s most highly cited
December 11, 2019
Seven researchers affiliated with CSHL are among the scientists producing the top 1 percent of the most highly-cited research in the world.
Making sense of the genome…at last
December 6, 2019
Quantitative biologists like Cold Spring Harbor Laboratory’s Adam Siepel are finally making sense of the flood of data contained in the human genome.
The Lab partners with award-winning magazine
December 6, 2019
Nautilus, an award-winning science magazine, has partnered with CSHL to bring the story of the lab’s scientists and research to a brand-new audience.
Research profile: Adam Siepel
November 12, 2019
Adam Siepel, Chair of the Simons Center for Quantitative Biology, uses advanced computational methods to solve complex biological questions.
A home like no other, Cold Spring Harbor Laboratory
November 7, 2019
Hear why our campus, our community, and our collaborative nature makes us a place that so many scientists call "home."
Scientists take action to prevent sexual harassment and bias
November 7, 2019
Scientists gathered at Banbury Center last year to discuss ways to prevent gender bias and sexual harassment in science.
Nobel laureate William Kaelin draws a crowd
October 29, 2019
Nobel laureate and cancer researcher William Kaelin holds a seminar on his award-winning research at CSHL.
Seeking better treatment for ALS, Lou Gehrig’s disease
October 29, 2019
Researchers found that ‘jumping genes’ were de-silenced in post-mortem tissue samples of ALS patients.
Peter Koo wants to understand how machines learn biology
September 20, 2019
Dr. Peter Koo joins the CSHL faculty as an assistant professor. His focus is on exploring how artificial intelligence integrates with biology and genomics.
Cancer drugs don’t always work as intended, researchers warn
September 11, 2019
Ten experimental cancer drugs kill tumors in ways that are entirely different than how clinicians thought they did, revealing important insights.
Event: Public Lecture: Seeing With Sequencing
August 8, 2019
Come hear from three quantitative biologists as they discuss how they see with sequencing to solve mysteries ranging from the genetics of evolution.
Sheltzer wins Presidential Early Career Award
July 9, 2019
CSHL Fellow Jason Sheltzer is a recipient of the Presidential Early Career Award for Scientists and Engineers for his work in cancer research.
Lee awarded Chan Zuckerberg Initiative Human Cell Atlas grant
June 21, 2019
Assistant Professor Je H. Lee has been named a recipient of one of the Seed Networks for the Human Cell Atlas to study breast tissue.
An essay from the President: Biology for the planet
May 16, 2019
CSHL plant scientists are looking for solutions to the biggest questions in agriculture as environments are reshaped by climate change.
Seeing with sequencing—A public lecture with three CSHL experts
April 19, 2019
Quantitative biologists discuss how physics, modern computing power, and a new perspective on biology can make sense of our complex genomes.
David McCandlish named Sloan Research Fellow
February 19, 2019
Assistant Professor David McCandlish has been named a 2019 Sloan Research Fellow for his promising work in the field of quantitative biology.
CSHL Fellow Jason Sheltzer wins innovation award
January 25, 2019
CSHL Fellow Jason Sheltzer wins the Damon Runyon-Rachleff Innovation Award for his work on cancer.
The year of CRISPR
December 26, 2018
A look at the various labs across CSHL that utilize CRISPR in their research, and the groundbreaking discoveries they help uncover.
How does natural selection affect the genome?
December 18, 2018
Adam Siepel explains how natural selection can tell researchers how informative sifting through the complex human genome will be.
How much are we learning? Natural selection is science’s best critic
December 17, 2018
Researchers determine that natural selection and our evolutionary history may be the best guides for future research.
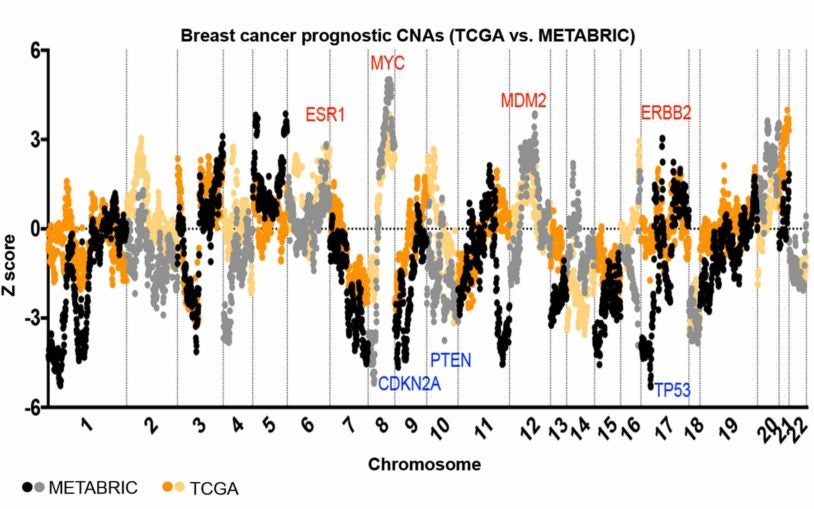
Taking uncertainty out of cancer prognosis
December 11, 2018
An analysis of 20,000 patients has revealed that copy number variations in specific gene sites can help predict how deadly a cancer will be.
Taking uncertainty out of cancer prognosis
December 11, 2018
CSHL Fellow Jason Sheltzer has analyzed nearly 20,000 cancer patient histories and genetic data to take the guesswork out of prognosis.
Molly Hammell wins CZI award for ALS study
December 5, 2018
Associate Professor Molly Hammell wins award for proposed study to find transposable elements that are implicated in ALS.
Base Pairs Episode 17: Genomes, justice, and the journey here
September 15, 2018
A look at how gene-mapping works, what scientists can tell by looking at your genome, and what it means for your privacy.
Genomes, justice, and the journey here
September 15, 2018
CSHL Professor W. Richard McCombie discusses genomic privacy and sequencing technology in this episode of Base Pairs.
Massive genome havoc in breast cancer is revealed
July 12, 2018
Researchers have made a highly detailed map of 20,000 structural variations in a cancer cell’s genome
Alexander Dobin dives into genomic data
June 8, 2018
Alexander Dobin joins the faculty as its newest assistant professor, working on the computational side of genomics research
A science writer’s quest to understand heredity
May 30, 2018
LabDish spoke with science writer Carl Zimmer about what he learned about heredity as he zig-zagged through CSHL while writing his new book.
Portrait of a Neuroscience Powerhouse
April 27, 2018
A relatively small neuroscience group at CSHL is having an outsized impact on a dynamic and highly competitive field
Base Pairs Episode 14.5: Medicine and mad scientists
April 16, 2018
A follow-up discussion with CSHL Fellow Jason Sheltzer from Base Pairs episode 14, “The cancer answer that wasn’t.”
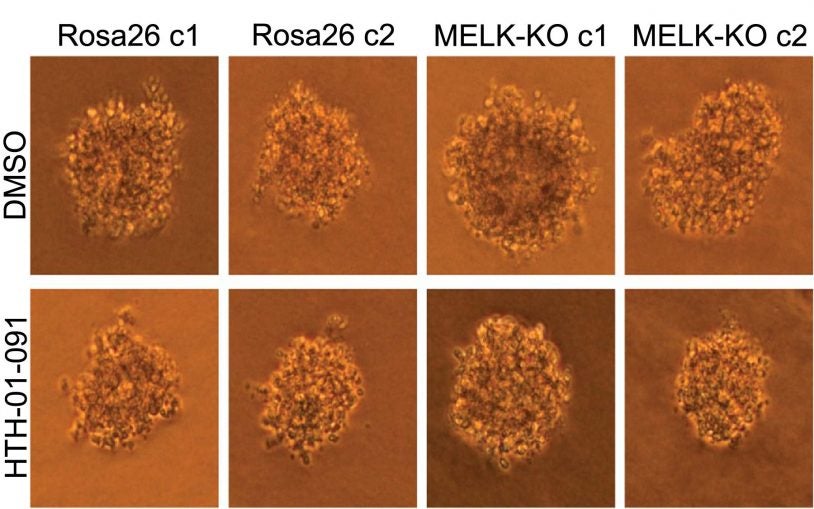
The cancer answer that wasn’t
March 15, 2018
We look at the "reproducibility crisis" in science, with a cancer researcher discovering something he didn't expect while experimenting with MELK
Base Pairs Episode 14: The cancer answer that wasn’t
March 15, 2018
We look at the "reproducibility crisis" in science, with a cancer researcher discovering something he didn't expect while experimenting with MELK.
Science self-corrects: cancer gene does not pass reproducibility test
February 13, 2018
CSHL Fellow Jason Sheltzer and his research team use CRISPR to discover that MELK is not actually involved in cancer.
Autism genetics study calls attention to impaired motor skills, general cognitive impairment
February 7, 2018
New research on the genetic causes of autism calls attention to diminished motor skills and suggests the importance of broad cognitive impairment
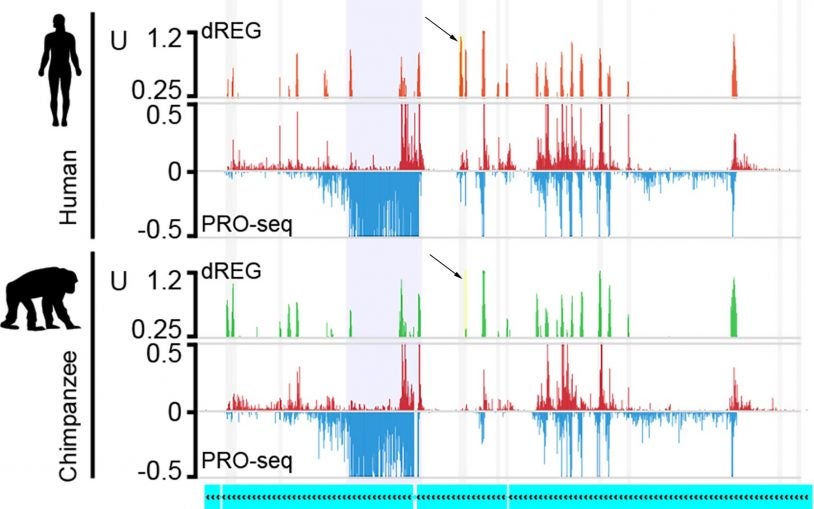
Evolving sets of gene regulators explain some of our differences from other primates
January 29, 2018
What makes us different from our primate relatives? Gene regulation is one important evolutionary factor
Base Pairs Episode 13: A lesson in class
December 15, 2017
We share three stories about classification in life sciences and how genetic information is changing how we define important categories.
A lesson in class
December 15, 2017
In this episode of Base Pairs, we discuss how genetic information is changing how we define important categories.
New method to determine before surgery which prostate tumors pose a lethal threat
December 1, 2017
The news about prostate cancer can be confusing. It’s the third most common cancer type among Americans.
Next-gen cancer test
November 24, 2017
Knowing that cancers become lethal when they spread, investigators at Cold Spring Harbor Laboratory seek a way of detecting tumors much earlier
CRISPR helps scientists find cancer’s (true) weaknesses
August 23, 2017
Jennifer Doudna (co-discoverer of the CRISPR gene editing tool) and CSHL's Jason Sheltzer talk breast cancer research.
New statistical method finds shared ancestral gene variants involved in autism’s cause
June 21, 2017
Researchers find children with autism are genetically more like other autistic children than their unaffected siblings.
Reconstructing ancient human history from DNA
June 20, 2017
Free public lecture featuring Adam Siepel, Ph.D., CSHL Professor and Chair of the Simons Center for Quantitative Biology.
Base Pairs Episode 9.5: Redefining biologists, redefining genes
May 16, 2017
We talk with Molly Hammell about peering into deep space using a high-tech telescope, we also talk to Tom Gingeras about redefining the gene.
Redefining biologists, redefining genes
May 16, 2017
Drs. Molly Hammell and Thomas Gingeras talk about redefining what a biologist is and what genes are.
Base Pairs Episode 9: Dark matter of the genome, part 2
April 15, 2017
In this episode of Base Pairs, we question the mythos that is “junk DNA” and explore the mysterious non-coding portions of the genome.
Dark matter of the genome, part 2
April 15, 2017
This episode of Base Pairs deals with the myth of "junk DNA" and why scientists are interested in non-coding portions of the genome.
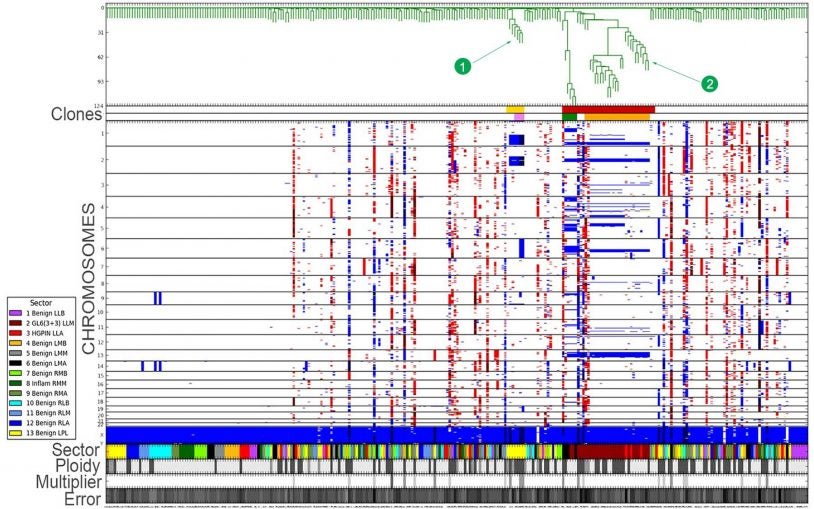
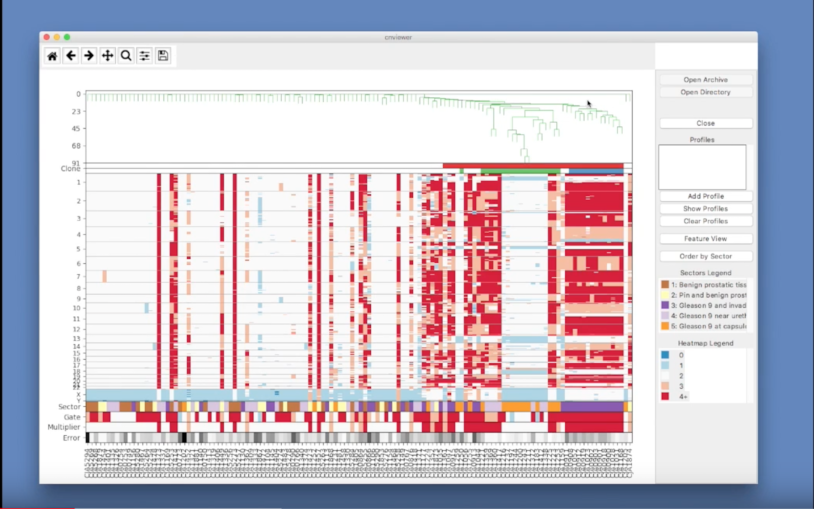
What is single-cell genome viewer?
March 31, 2017
Single-cell genome viewer is a graphical interface, integrating single-cell genomics and conventional pathology of tumors.
Research suggests a possible role for a storm of ‘jumping genes’ in ALS
March 27, 2017
New research demonstrates that some transposons are no longer effectively inhibited, resulting in a storm of jumping genes, leading to DNA damage.
What a real-life science test looks like
March 24, 2017
By revealing evidence that contradicts the rationale for a new cancer drug, a pair of student scientists learns firsthand that when you do science.
Dark matter of the genome, part 1
March 15, 2017
This episode of Base Pairs digs into "dark matter" a type of genetic information that could help scientists better understand diseases like
Base Pairs Bonus Episode: Molecules and a mission
January 27, 2017
In this bonus episode, we explain the molecules and the metaphor of our podcast name: Base Pairs.
 Building publication list.
Building publication list.
Semir Beyaz
Are you really what you eat? Our goal is to uncover the precise mechanisms that link nutrition to organismal health and disease states at the cellular and molecular level. A particular focus in our lab is to understand how dietary perturbations affect the immune system and contribute to the risk of diseases that are associated with immune dysfunction such as cancer.
Nyasha Chambwe
My research focuses on identifying the genetic and molecular features of cancers that differ across racial and ethnic groups, and the extent to which these differences reveal or explain race and ethnicity-based cancer health disparities.

Kenneth Chang
RNA interference (RNAi) and CRISPR are widely used to functionally investigate mammalian genomes. It is our goal to develop and optimize these gene perturbation platforms to improve their effectiveness in understanding the biology of diseases.

Camila dos Santos
Among the changes that occur during pregnancy, those affecting the breasts have been found to subsequently modify breast cancer risk. My laboratory investigates how the signals present during pregnancy permanently alter the way gene expression is controlled and how these changes affect normal and malignant mammary development.

Douglas Fearon
I’m studying how to harness the power of the immune system to fight cancer. Our underlying premise is that the microenvironment within a tumor suppresses the immune system. We have found a way to eliminate this suppression in the mouse model of pancreatic cancer, which has led to development of a drug for human pancreatic cancer that will enter phase 1 clinical trials in 2015.

Sara Goodwin
I work on adapting and developing new methods/techniques for genome and transcriptome sequencing.

Tobias Janowitz
Cancer is a systemic disease. Using both laboratory and clinical research, my group investigates the connections between metabolism, endocrinology, and immunology to discover how the body’s response to a tumor can be used to improve treatment for patients with cancer.

Alexander Krasnitz
Many types of cancer display bewildering intra-tumor heterogeneity on a cellular and molecular level, with aggressive malignant cell populations found alongside normal tissue and infiltrating immune cells. I am developing mathematical and statistical tools to disentangle tumor cell population structure, enabling an earlier and more accurate diagnosis of the disease and better-informed clinical decisions.

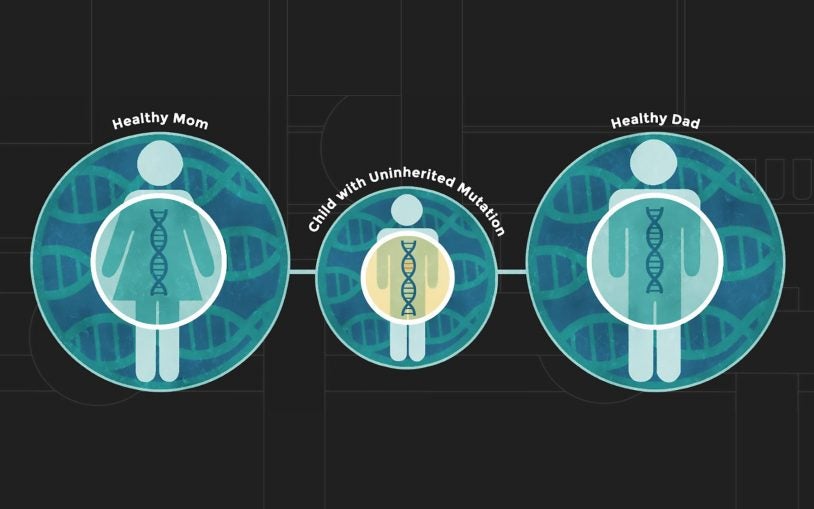
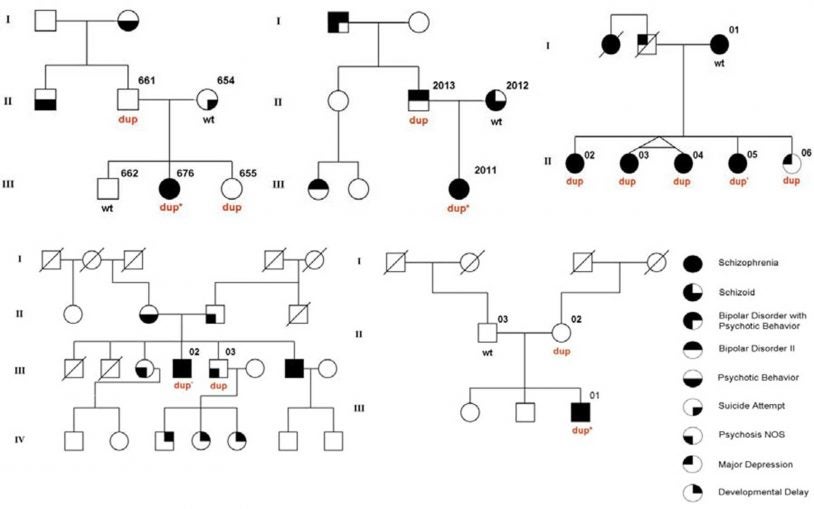
Dan Levy
We have recently come to appreciate that many unrelated diseases, such as autism, congenital heart disease and cancer, are derived from rare and unique mutations, many of which are not inherited but instead occur spontaneously. I am generating algorithms to analyze massive datasets comprising thousands of affected families to identify disease-causing mutations.

W. Richard McCombie
Over the last two decades, revolutionary improvements in DNA sequencing technology have made it faster, more accurate, and much cheaper. We are now able to sequence up to 10 trillion DNA letters in just one month. I harness these technological advancements to assemble genomes for a variety of organisms and probe the genetic basis of neurological disorders, including autism and schizophrenia, better understand cancer progression and understand the complex structures of the genomes of higher plants.

Hannah Meyer
A properly functioning immune system must be able to recognize diseased cells and foreign invaders among the multitude of healthy cells in the body. This ability is essential to both prevent autoimmune diseases and fight infections and cancer. We study how a specific type of immune cells, known as T cells, are educated to make this distinction during development.

Adam Siepel
I am a computer scientist who is fascinated by the challenge of making sense of vast quantities of genetic data. My research group focuses in particular on questions involving molecular evolution and transcriptional regulation, with applications to cancer and other diseases as well as to plant breeding and agriculture.

Chris Vakoc
Cancer cells achieve their pathogenicity by changing which genes are on and off. To maintain these changes in gene expression, cancer cells rely on proteins that interact with DNA or modify chromatin. My group investigates how such factors sustain the aberrant capabilities of cancer cells, thereby identifying new therapeutic targets.

Peter Westcott
The mutational processes that drive cancer also expose it to the immune system. Therapies that invigorate anticancer immunity can be astonishingly effective, but only in a subset of patients. We are developing powerful new strategies to study how the immune system and cancer coevolve, with the goal of expanding the curative potential of immunotherapy to more patients.

Michael Wigler
Devastating diseases like cancer and autism can be caused by spontaneous changes to our DNA—mutations first appearing in the child, or in our tissues as we age. We are developing methods to discover these changes in individuals, tumors, and even single cells, to promote early detection and treatments