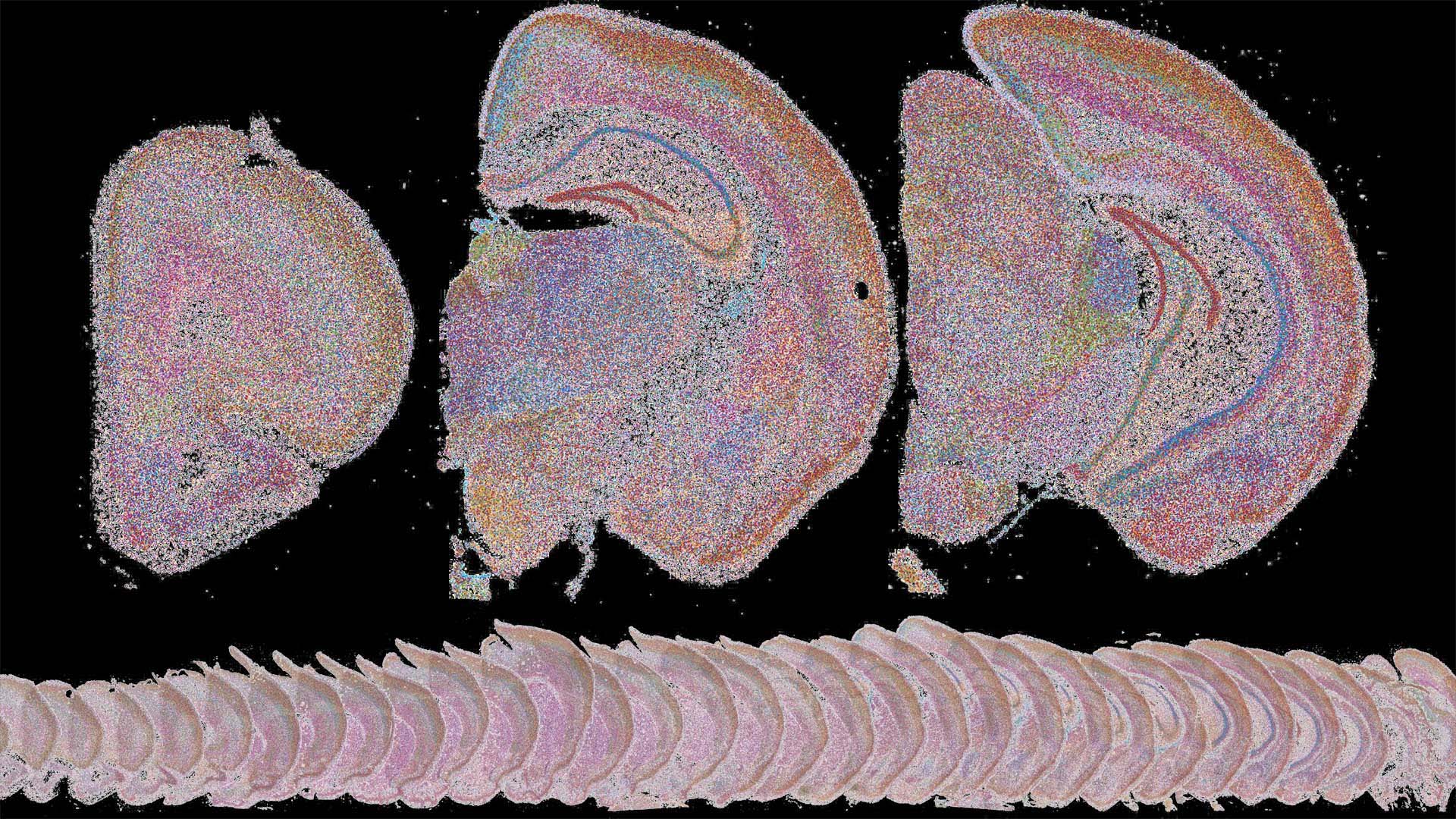
Understanding the connections between different brain areas could pave the way to better treatment strategies for conditions like Alzheimer’s, schizophrenia, and depression.
In 2019, as a postdoc in Cold Spring Harbor Laboratory’s (CSHL’s) Zador lab, Xiaoyin Chen helped develop a technique to map these connections. BARseq identifies cells in the brain by the genes they use and traces the connecting neural circuitry. Early versions of BARseq mapped gene expression across thousands of neural pathways, using “barcodes” or short snippets of RNA.
Chen is now an assistant investigator at the Allen Brain Institute. He recently reunited with CSHL Professor Anthony Zador to upgrade BARseq’s capabilities. What does that look like? Instead of thousands of neurons, BARseq can now map millions. Chen says:
“We are focused on pushing BARseq forward. We want to make this easy for everybody to use, faster, more sensitive. Can we read out more information with it? With much higher scale, you can start to answer different questions.”
The team began their search for answers in the brain’s visual cortex. Sight is one of the most common ways humans perceive the world. Information travels from the eyes to the visual cortex for processing. But what happens in the brain when the visual cortex’s neural inroads are cut or don’t form at all?
“People have known for a while that visual inputs are very important in shaping the brain,” Chen explains. “But we don’t know, at the exact cell-type resolution BARseq provides, what actually happens.”
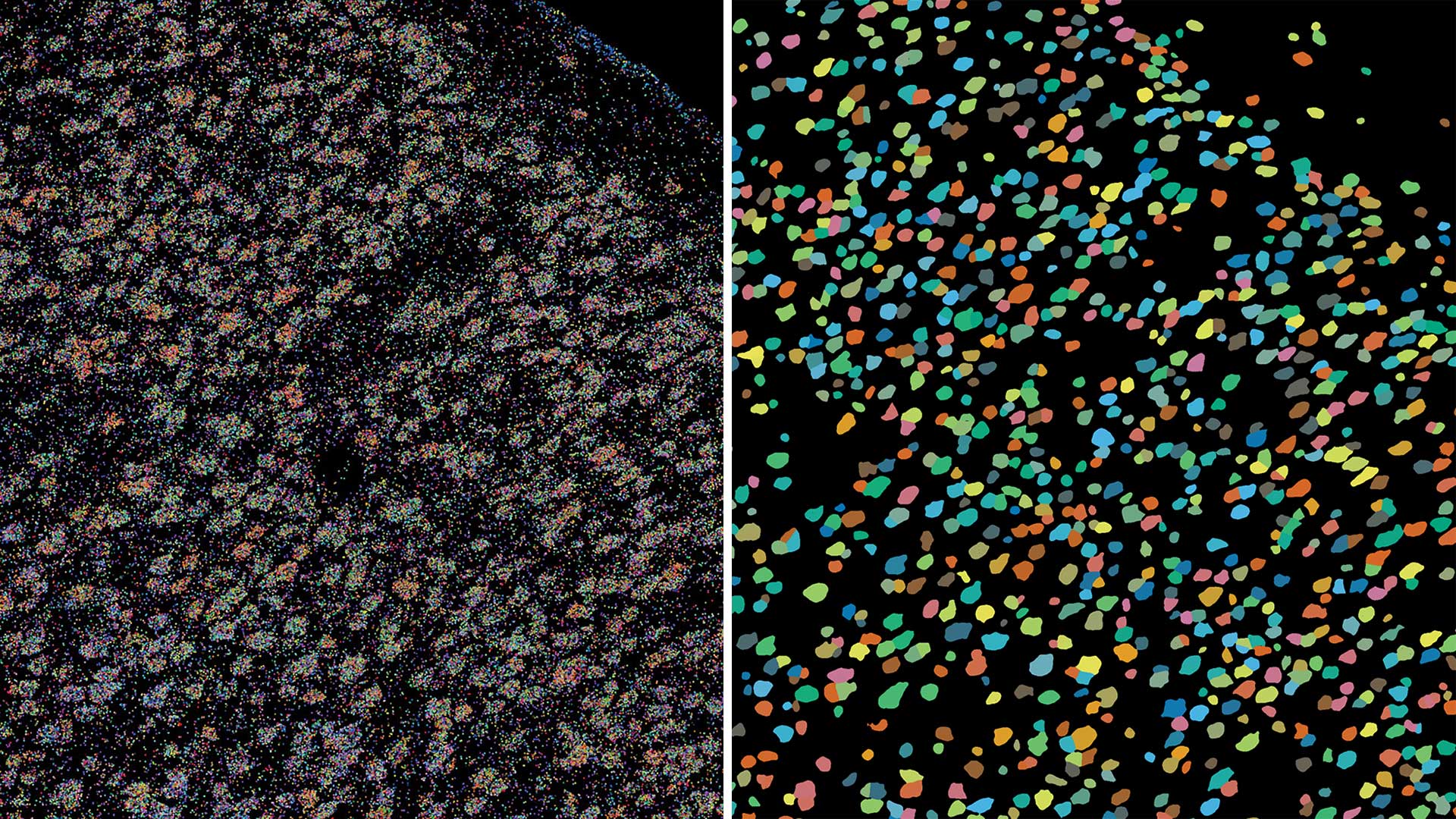
The team used BARseq to map the brains of nine mice and traced gene expression in each mouse’s visual cortex. It’s the first time the technique has been used to map this many entire brains. Amazingly, the team found that if the mice went blind, the genes in the visual cortex started to look like those in neighboring cortical areas of the brain.
“The effects of losing vision were very broad,” Chen explains. “The visual cortex itself changes. It becomes more similar to the areas around it. There are still a lot of questions about how development controls this patterning.”
Chen is now working to expand BARseq’s capabilities even further. He and his team are using the technique to investigate how connections are wired in developing brains and how these connections evolve.
“Understanding how cortical areas are set up is the first step in understanding these connections,” he says. “But it’s not enough. We still need to discover how they progress during development. BARseq can bring us closer to that goal.”
Written by: Nick Wurm, Communications Specialist | wurm@cshl.edu | 516-367-5940
Funding
U.S. Department of Health & Human Services, National Institutes of Health, Brain Research Foundation, Intelligence Advanced Research Projects Activity, Paul G. Allen Frontiers Group
Citation
Chen, X., et al., “Whole-cortex in situ sequencing reveals input-dependent area identity”, Nature, April 24, 2024. DOI: 10.1038/s41586-024-07221-6
Principal Investigator

Anthony Zador
Professor
The Alle Davis and Maxine Harrison Professor of Neurosciences
M.D., Ph.D., Yale University, 1994